Dive Deep into Multi-omics
Explore, Visualize, and Analyze Multi-omics Data with our Smart Software Solutions!
Dive into Multi-omics Data
We created an extensive collection of easy-to-use tools to visualize and analyze multi-omics data, from peak picking to concentrations, from normalization to metabolic pathway analysis
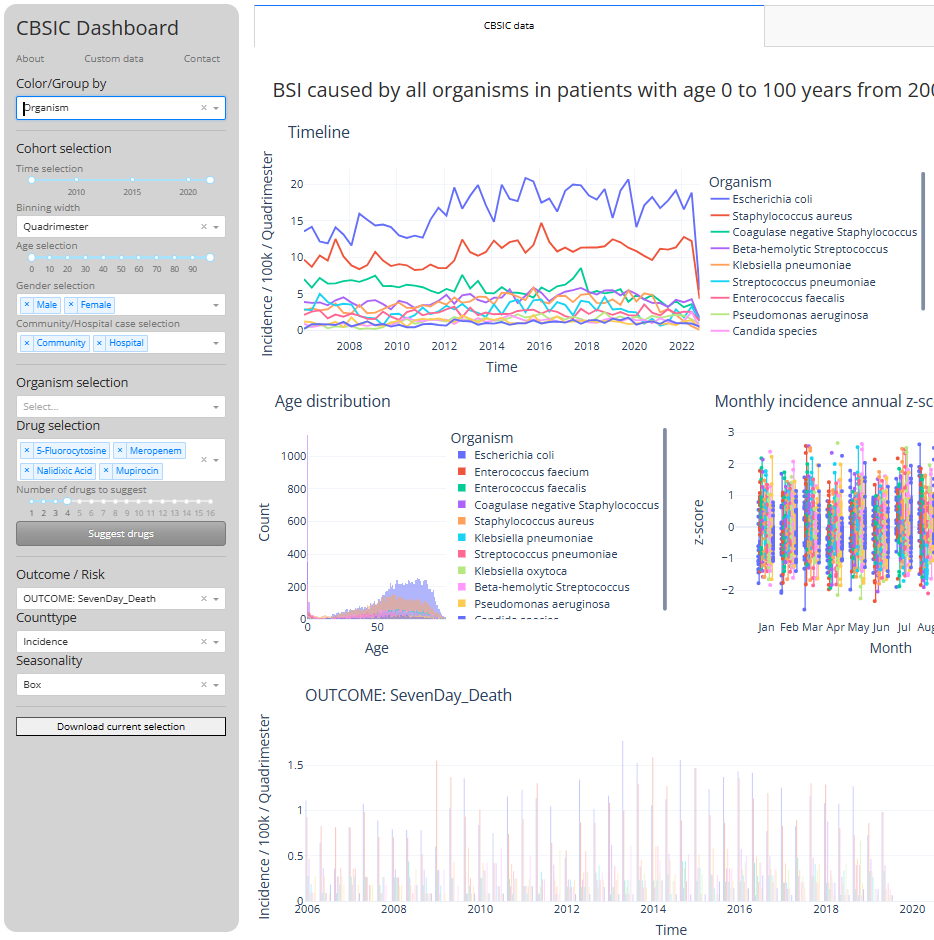
The Calgary BSI Cohort dashboard allows easy access to detailed epidemiological data from CBSIC data such as patient age category, sex, organism names, acquisition type, hospital, seasonal distribution of infections, outcomes and antimicrobial resistances
The ProteomicsQC enables automatic processing of Quantitative Proteomics LC-MSMS data, along with visualization of 69 quality control metrics and ML-driven outlier analysis
Metabolomics Integrator peak integration (MINT) is a post-processing tool for liquid chromatography-mass spectrometry (LCMS) based metabolomics
Covid detection using proteomics
SCALIR is used to standardize metabolomics data by using standard samples and incorporates a novel algorithm for detecting linear ranges in a set of (x,y) points covering several orders of magnitude
Metabolic Interactive Nodular Network for Omics (MINNO) is a D3 javascript library-based web application to visualize biological data onto metabolic networks. This tool offers various ways to share data and results to facilitate collaboration across multiple research groups globally
Filtering Utility for Grouping Untargeted Mass Spectrometry (FUGU-MS) datasets is a shiny application for untargeted analysis of metabolomic data
GWAS tool
GWAS tool for Genome wide association studies
Phi-values
Phi-values tool computes de Phi-correlation coefficients for a set of binary outcome variables in the CBSIC
Genomic processing
Analysis and annotation of microbial DNA sequencing data using the most up-to-date tools and databases